Enter the Entrez ID, RefSeq ID, or sequence for your gene of interest in
FASTA format into the box on the home page.
FASTA format requires two lines per entry. The first line for an entry must begin with a greater-than (">") symbol and the second line must have either a single Entrez ID, a single
RefSeq ID, or a sequence of ATGC or N. Blank lines are not permitted. Multiple entries may be submitted at once on separate lines.
If Entrez IDs are used as input, hg38 is used as the human reference and mm10 is used as the mouse reference. If RefSeq IDs are used as input, the most recent version of the sequence is
pulled from NCBI.
Advanced Parameters
| Predictions per gene | The number of predictions to return for each FASTA entry. Default:5 |
| Remove constructs with EcoRI or Xho sites | By default, sequences containing an EcoRI or XhoI restriction site (GAATTC or CTCGAG)
will be removed from the predictions. To include them, uncheck this box. |
| Sequence before most proximal poly-A site (for Entrez IDs) | Consider only the gene sequence before the most proximal annotated polyadenylation site for shRNA candidates.
This option is enabled by default and only applies to predictions using Entrez IDs. |
| Coding sequences only (for Entrez IDs) | Remove 5'UTR and 3'UTR sequences from predictions when using an Entrez ID as input. |
| Show predictions present multiple times in the same gene | Include shRNAs predicted to hit the target gene in multiple locations. By default these sequences are excluded. |
| Show predictions present in other genes (off-targets) | Include shRNAs targeting a sequence appearing in another mouse or human gene. By default these sequences are excluded. |
To convert a list of gene names or gene IDs to Entrez IDs, use the
Gene ID Conversion tool from DAVID. Copy-paste your
list of genes into the box on the left or upload a file with your gene IDs. Next, choose the type of gene identifier used in your list or choose "Not Sure" if you don't know. Choose
"Gene List" as your list type. Next, using "Option 1", choose ENTREZ_GENE_ID as your goal identifier and click "Submit to Conversion Tool."
NCBI Information
If either a RefSeq or Entrez ID is entered as input, a link to the relevant NCBI page is displayed.
Sequence of intersection
If the Entrez ID of a gene is entered as input, shRNAs are predicted for sequences present in all isoforms of that gene. A figure showing all isoforms for a gene along with the
shared sequences is provided with the results. Additionally, the sequence of the shared isoforms is provided. An asterisk (*) is used to separate exon junctions.
shRNAs will not be predicted across these junctions.
Intersection Figure
If the Entrez ID of a gene is entered as input, a figure will be generated showing all protein-coding isoforms of that gene along with the regions of the gene that are present
in all isoforms (the intersection). 5'UTR and 3'UTR sequences are in light blue, coding sequences (CDS) are in blue, and the intersection sequence is in dark blue. The bar to
the left of the figure can be used to zoom in/out of the figure. Red bars below the intersection sequence indicate the predicted targeting position of the shRNA in the table
below.
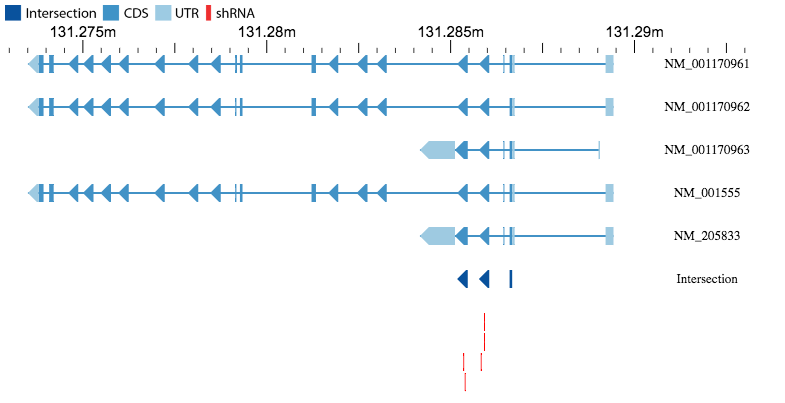
Moving your cursor over one of these shRNA elements will show the label of the shRNA.

Results Table
A results table will be generated for each entry and shows the predicted shRNAs sorted by their splashRNA score.
| Label | Unique shRNA name defined by the feature name and the position of the shRNA |
| Antisense Guide Sequence | sequence of the shRNA guide sequence. |
BLAST Symbol  | BLAST the shRNA sequence against NCBI's non-redundant database. |
| SplashRNA | splashRNA score for the shRNA |
| Warnings | list of warnings for this shRNA (see below for more information) |
| Mouse matches | Number of mouse genes with perfect 22mer matches to this shRNA (only appears if sequence is non-unique). Hovering over this will show the list of matching entrezIDs. |
| Human matches | Number of mouse genes with perfect 22mer matches to this shRNA (only appears if sequence is non-unique). Hovering over this will show the list of matching entrezIDs. |
Download
Results can be downloaded as a CSV (comma-separated value file) by clicking the "Download" button at the bottom of the page. The downloaded file will contain all of the information
in the results tables as well as the full 97-mer construct for each predicted shRNA. The layout of the file is as follows:
| Feature | Name of query |
| shRNA.name | Unique shRNA name defined by the feature name and the position of the shRNA |
| Antisense.Guide.Sequence | sequence of the shRNA guide sequence |
| SplashRNA | splashRNA score for the shRNA |
| 97mer.construct | full 97mer construct containing the shRNA |
| Warnings | list of warnings for this shRNA (see below for more information) |
| Mouse.22mer.match.genes | Number of mouse genes with perfect 22mer matches to this shRNA |
| Human.22mer.match.genes | Number of human genes with perfect 22mer matches to this shRNA |
| Mouse.22mer.match.entrezIDs | List of matching mouse entrez IDs |
| Human.22mer.match.entrezIDs | List of matching human entrez IDs |
Warnings and Errors
- Low score - Strong shRNAs generally have a splashRNA score above 1.0. Very strong shRNAs generally have scores above 2. Low scores can be due to a short input
sequence or gene, if only a small portion of a sequence is present in all isoforms, or from a very GC-rich sequence. Increasing the length of the sequence or choosing
a single isoform (using a RefSeq ID) may help increase the scores of the predicted shRNAs.
-
Short intersection - This warning occurs if the length of the sequence shared between all isoforms of a gene is less than 60% of the full gene length and
the scores of the predicted shRNAs are low. In most cases, choosing the isoform that is most relevant to you will improve results.
-
EcoRI or Xho site in 97mer construct - Either an EcoRI (GAATTC) or Xho (CTCGAG) restriction site is present in the 97mer construct.
-
Your sequence contains Ns - shRNAs can only be predicted over sequences where all nucleotides (22bp) are A, G, T, or C. Predictions will only be made on 22mers
that do not contain Ns.
-
No intersection - When an Entrez ID is entered as input, predictions are made on the sequences that are present in all isoforms of that gene. In some cases, there
are no sequences that are shared across all isoforms. In this case, it is best to choose the isoform that is most relevant to your system and enter the RefSeq ID for that isoform.
-
FASTA file not recognized - The input was likely not in FASTA format. FASTA format requires two lines per entry.
The first line for an entry must begin with a > and the second line must have either a single Entrez ID, a single RefSeq ID, or a sequence of ATGC or N. Blank lines are not permitted.
You may also get this error if a character other than A, T, G, C, or N is in your sequence. Lines beginning with a # will be ignored. See the home page for an example.

